Description
| Application: | In vitro transcription, enzyme assay For research use only! |
| Identity: | Bacteriophage T7 enzyme, recombinant from yeast (Hansenula polymorpha) |
| Unit definition: | The amount of enzyme required to hydrolyse 4 nmol of NTP per hour at 37 °C in an in-vitro transcription reaction using 1 µg of T7 template encoding firefly luciferase. Released diphosphate is measured as phosphate using iPPase and the Malachite Green reaction. |
| Size: | 889 amino acids; MWcalc. = 99,7 kDa (monomer) |
| Purity: | > 95 % by SDS-PAGE |
| Buffer composition: | 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1 % v/v Tween20, 50 % v/v glycerol, 20 mM Tris-HCl, pH 7.5 (@ 25 °C) |
| Storage: | store at -20 °C, avoid freeze/thaw cycles |
| Expression system: | Hansenula polymorpha (yeast) |
| Composition: | Animal component free |
Additional information:
T7 RNA Polymerase ( T7 RNAP, EC 2.7.7.6) is a DNA-directed RNA polymerase cloned from bacteriophage T7 and overexpressed in yeast H. polymorpha. The enzyme is active as monomer and shows high specificity for its genuine promoter sequence. Except Mg 2+ no other co-factor is required for the polymerase reaction. The reaction is driven by the hydrolysis of the phosphate anhydride bond of the NTP. The enzyme is a preferred choice for in vitro transcription. iPPase is frequently employed in vitro to hydrolyse diphosphate liberated by the RNA polymerase reaction (related product).
Coomassie stained SDS-PAA Gel
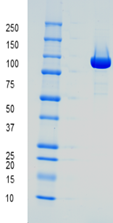
Lane 1: molecular weight marker; Lane 2: buffer; Lane 3: ARTES T7 RNAP, better 95 % purity
Agarose gel of samples of IVT reactions
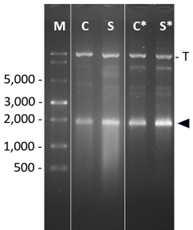
Lane M: ss RNA marker; Lane C: T7 RNAP comparator; Lane S: ARTES T7 RNAP, Lanes C* and S*: same as before, but IVT in presence of ARTES iPPase
The arrow head marks the transcript band of about 1,900 nucleotides and T the template DNA band.
If you need any more information at all, or if you’ve cited our amazing T7 Polymerase in a publication, please don’t hesitate to get in touch! We would absolutely love to hear from you and would be thrilled to reward your research.